Root-Project: Introducing SOFIE
Fast Inference of Deep Learning models following the ONNX Standards
The Toolkit for Multivariate Analysis (TMVA) is a sub-module of the Root-Project which provides a machine learning environment to undertake the processing and evaluation of multivariate classification and regression techniques, primarily used in High-energy Physics research. Due to the bulk size of data (approximately in petabytes) being generated daily from various research activities and the demand for quick predictions for faster results, it has become highly necessary to have a fast inference engine for the trained machine learning models for the frequent use of trained machine learning models.
The TMVA Team built a tool called SOFIE, which stands for System for Optimized Fast Inference code Emit . SOFIE is a fast inference system, having its own intermediate representation of the deep learning models following the ONNX standards. With SOFIE, the TMVA team aims to achieve easier and faster deployments of trained machine learning models in a production environment.
My first primary task in my project for Google Summer of Code was to develop the capability to store and read this intermediate representation, thus making it serialisable.
In my third blog of this series, I will be introducing SOFIE and how I worked on making it serialisable.
Table of Contents
1. About SOFIE
SOFIE is an ONNX operator-based infrastructure that provides the capability for having an intermediate representation of a trained deep learning model and generates an inference code for the prediction of output based on some provided input. At its core, it generates C++ functions that are easily invokable for fast inference. In its basic usage, it takes an ONNX file as input, and after parsing it, generates a .hxx C++ header file having a infer function that can be easily included where the model is supposed to be used.
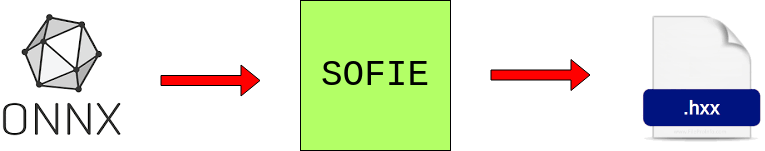
Interface
SOFIE has some pre-requisites which include BLAS/Eigen for executing the generated inference code, and Protobuf3, for parsing the ONNX files.
For example usage, SOFIE has the following interface for operation
using namespace TMVA::Experimental;
SOFIE::RModelParser_ONNX Parser;
SOFIE::RModel model = Parser.Parse(“./example_model.onnx”);
model.Generate();
model.OutputGenerated(“./example_output.hxx”);
The above code can be executed in the ROOT Command line for having an understanding of how SOFIE works. Executing these will generate a .hxx header file which will contain the inference code for the model.
Architecture
SOFIE contains two major classes, which include RModel and ROperator. RModel is the primary class that holds the essential model information, and ROperator is the abstract base class from which various operators are derived. Following ONNX standards, the ROperators are responsible for generating specific inference code to operate on input tensors and provide the outputs as per attributes provided.
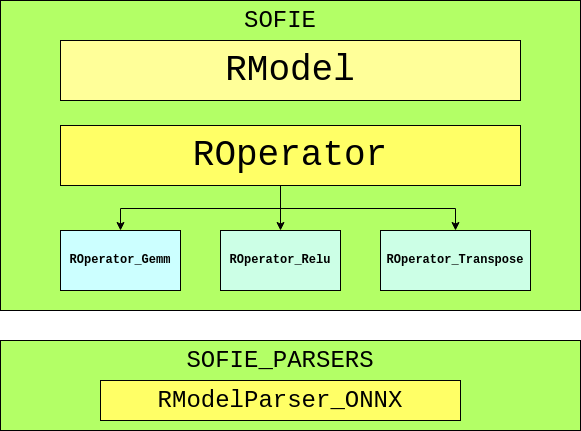
Another sub-directory under TMVA was developed called sofie-parsers to hold the ONNX parser named as, RModelParser_ONNX which basically parses an ONNX file and loads the read value into an RModel object, and then returns the object. The reason for keeping the ONNX parser separate from the directory where RModel and ROperators reside is because of Protobuf. SOFIE was found crashing while using any other package which also uses Protobuf because of compatibility issues. MethodPyKeras present in PyMVA is one such file because TensorFlow (Back-end for Keras) was seen having compatibility issues with Protobuf while it was used by SOFIE.
An example header file generated by SOFIE after parsing an ONNX file will look like,
//Code generated automatically by TMVA for Inference of Model file [model.pt] at [Sat Jul 10 22:41:27 2021]
#include<algorithm>
#include<vector>
namespace TMVA_SOFIE_model{
namespace BLAS{
extern "C" void sgemm_(const char * transa, const char * transb, const int * m, const int * n, const int * k,
const float * alpha, const float * A, const int * lda, const float * B, const int * ldb,
const float * beta, float * C, const int * ldc);
}//BLAS
float tensor_fc1weight[120] = {0.789854884, 0.376688957, 0.100161314, -0.0823104382, 0.397924304, 0.304954529, -0.617295027, 0.0845124722, -0.145524979, 0.671475768, -0.224907994, 0.11630702, 0.0874925852, 0.926566958, 0.764389873, -0.816925645, 0.138820171, 0.446122766, -0.751768351, -0.628336191, 0.65862155, 0.255001545, -0.620595455, 0.034793973, -0.877461433, 0.560568094, 0.000694513321, -0.360476613, -0.4559623, -0.89475286, -0.318023324, -0.672464848, -0.58325696, -0.874113441, 0.972242355, -0.655378938, 0.10801971, 0.863679409, 0.546122909, -0.595589399, 0.942520261, -0.0177378654, -0.879495144, 0.579633832, 0.78146708, -0.606159925, 0.0977220535, 0.106507063, 0.042394042, -0.620597243, 0.165384531, 0.98827672, 0.331049085, 0.0309733152, 0.532483697, -0.647528291, 0.777747393, -0.432416439, 0.280081153, 0.778528214, 0.562044621, 0.892722726, -0.738910079, -0.0454969406, 0.618175983, 0.0212159157, 0.850445628, 0.0888565779, -0.733002305, -0.816845894, 0.36951673, -0.861773133, -0.514285088, -0.745126009, -0.836292148, -0.193939567, 0.724980474, 0.332978487, -0.136577129, -0.431463122, -0.837541223, -0.491315842, 0.344886184, -0.494451523, -0.0386657715, -0.0283970833, -0.301973939, -0.0306991339, 0.964098096, 0.0289236307, 0.196783185, 0.531594157, -0.824845672, 0.188128829, -0.751537561, 0.764263868, 0.978678823, -0.494144678, -0.0894714594, 0.309134603, 0.974077702, 0.298210502, 0.0480248928, 0.118162513, -0.880085468, -0.937008262, -0.119312763, -0.126730084, 0.794969797, 0.385711908, 0.603180885, 0.949148774, -0.443132043, 0.428644657, 0.515355825, 0.100105286, -0.980962157, -0.610996366, -0.128770351, 0.737880945};
float tensor_fc1bias[120] = {0.156999707, 0.925517797, -0.82491076, -0.327555299, 0.300433517, 0.0843735933, 0.937375665, 0.90215981, 0.525184274, -0.388632417, -0.958357334, 0.691158652, 0.288623691, 0.317767382, 0.690718174, -0.108947277, 0.0974274874, -0.178514361, 0.22222352, 0.481612802, -0.725877881, -0.917040825, 0.401632786, 0.173210263, -0.807639718, 0.071665287, 0.926479101, 0.226339459, 0.912018657, -0.177851915, 0.0912634134, 0.457497716, -0.00866019726, 0.876882434, 0.967141509, -0.389477611, -0.397746086, 0.38465035, -0.0337526798, 0.598025203, -0.197502494, 0.985955477, -0.389883399, 0.280894637, -0.342840672, 0.280558348, -0.0282634497, -0.552011251, 0.416986227, 0.804085135, 0.745562077, 0.805344701, 0.0866900682, 0.903210402, 0.526807904, -0.492496252, -0.689859748, 0.411614537, 0.734182239, -0.385229945, -0.276290894, 0.853501678, -0.22111702, 0.00479888916, -0.631456971, 0.109048486, -0.0367674828, 0.465072155, 0.242459297, -0.775521278, -0.864489079, 0.82010138, 0.96387732, 0.428108692, 0.301132321, -0.600532174, -0.824067235, -0.761821628, -0.717110753, -0.51265502, -0.4664675, -0.0190160275, 0.903397322, -0.107425809, 0.494693398, -0.48946774, -0.165202618, 0.74544704, 0.329532743, -0.564413428, 0.466064095, 0.998960733, 0.763867974, 0.569300771, -0.0771077871, 0.185100675, 0.799941301, -0.00554811954, 0.445079327, 0.255458713, -0.476707339, -0.519067168, -0.678400636, 0.508345485, -0.0559206009, 0.16065681, 0.369853497, -0.924479008, -0.525777817, -0.0643764734, 0.545757532, -0.754187346, -0.213796258, 0.662986755, -0.905717492, -0.338911414, 0.676254153, 0.315755367, 0.61975944, -0.942272425};
float tensor_4[14400];
float tensor_3[14400];
std::vector<float> infer(float* tensor_x1){
char op_0_transA = 'n';
char op_0_transB = 't';
int op_0_m = 120;
int op_0_n = 120;
int op_0_k = 1;
float op_0_alpha = 1;
float op_0_beta = 1;
int op_0_lda = 1;
int op_0_ldb = 1;
std::copy(tensor_fc1bias, tensor_fc1bias + 120, tensor_3);
BLAS::sgemm_(&op_0_transB, &op_0_transA, &op_0_n, &op_0_m, &op_0_k, &op_0_alpha, tensor_fc1weight, &op_0_ldb, tensor_x1, &op_0_lda, &op_0_beta, tensor_3, &op_0_n);
for (int id = 0; id < 14400 ; id++){
tensor_4[id] = ((tensor_3[id] > 0 )? tensor_3[id] : 0);
}
std::vector<float> ret (tensor_4, tensor_4 + sizeof(tensor_4) / sizeof(tensor_4[0]));
return ret;
}
} //TMVA_SOFIE_model
The infer function present here is used for inference on provided input tensor to predict outputs.
API Reference of SOFIE generated by DOxygen can be used to know more about the implementation and usage of the functions, classes and other data structures.
2. What are ROOT files?
ROOT file is like a UNIX file structure, which is stored in a machine-independent format containing directories and objects organized in an unlimited number of levels. Primarily used in Root-Project and TMVA, they have the functionality of serialisation, thus are used to save and read objects, hence making them persistent.
A .root file is created/handled using a TFile object.
TFile *file = new TFile("file.root","OPTIONS");
TFile file("file.root","OPTIONS");
where the OPTIONS can be one among CREATE, NEW, RECREATE, UPDATE, READ. Once created, this becomes the default for I/O and all file related I/O are executed on the initialized file. ROOT uses a compression algorithm based on the gzip algorithm supporting nine levels of compression, where the default is 1. The ROOT I/O is developed for fast and highly compressed read/write of data from the .root file.
Writing into a .root file
Having a serialisable class, we can write its object into a .root file.
//For a class exampleClass
TFile file("file.root","CREATE");
exampleClass object;
object.getData(); //To populate the object with some data
object.Write("object");
file.Close();
While calling the Write() function, a key needs to be passed which maps the object within the file. While reading the object back, the key is required, as a .root file can contain various objects and data structures.
Reading from a .root file
Having defined the class to be used to load an object from a file, we can read from .root file .
//For a class exampleClass
TFile file("file.root","READ");
exampleClass *objectPtr;
file.GetObject("object",objectPtr);
file.Close();
For more information on ROOT files, one may visit the official ROOT's Manual on ROOT Files from CERN here.
3. Serialising SOFIE
To make a class serialisable in ROOT, it's required to be inherited publicly from the TObject class. Inheriting TObject, provides the class with a Streamer() function which writes the current values of the data members by decomposing the object into its data elements, and in a similar way reading back the serialised data elements into an object of the class. The methods of the class are not written to the ROOT file. The Streamer() moves up calling all the streaming functions of the parent classes to fill the buffer, which is then written into the .root file. To ensure a class has a Streamer(), the ClassDef() macro needs to be called in the class declaration so that a default streamer is automatically generated.
//Default Streamer for class exampleClass
void Event::Streamer(TBuffer &R__b){
if (R__b.IsReading()) {
Event::Class()->ReadBuffer(R__b, this);
}
else {
Event::Class()->WriteBuffer(R__b, this);
}
}
While making the RModel class serialisable to store the model information into a .root file, it was required to develop a custom streamer for the class as it contains some complex data structure. The involved classes and structures were made serialisable by linking them in the LinkDef.h file with the appropriate #pragma link so that their underlying data structures can be found while streaming. The custom streamer was built majorly for making a structure capable of serialisable called TMVA::Experimental::SOFIE::InitializedTensor as it contains a data-member called fData which is declared as a shared_ptr. ROOT is unable to serialise the values pointed by a shared_ptr<void> and thus, the structure was modified to have a char* for pointing to the memory held by the shared_ptr. Appropriate functions are developed to convert the shared pointer to the persistent pointer and vice-versa. The developed functions were called in the Custom Streamer during the reading and writing of the .root file.
void RModel::Streamer(TBuffer &R__b){
if (R__b.IsReading()) {
RModel::Class()->ReadBuffer(R__b, this);
for(auto i=RModel::fInitializedTensors.begin(); i!=RModel::fInitializedTensors.end();++i){
i->second.CastPersistentToShared();
}
}
else {
for(auto i=RModel::fInitializedTensors.begin(); i!=RModel::fInitializedTensors.end();++i){
i->second.CastSharedToPersistent();
}
RModel::Class()->WriteBuffer(R__b, this);
}
}
The development of Serialisation of RModel class is tracked with PR#8666
With this, we complete serialising the RModel class. This will be used to store the model information into .root files. Storing the model into .root file will make serializing the model configuration and weights into a compressed file which can be read to generate the inference code later.
In my next blog, I will be introducing the RModelParser_Keras, developed to translate any Keras model into an RModel object, so that the fast inference code for a Keras .h5 file can be generated from the parsed RModel.
annyeong,
Sanjiban
