GSoC'21: Community Bonding Period
Interacting with the community at CERN and starting off my work
Soon the results for GSoC'21 were published, the community bonding period kicked off starting the 17th of May'2021 till the 7th of June'2021. During this time, the participants are supposed to communicate with their respective mentors and organization administrators discussing their project goals, finalizing the proposed plan, and familiarising themselves with the community practices to establish a working strategy.
In my second blog in GSoC with the CERN-HSF series, here I present my experience during the Community Bonding period, where I will be describing my interaction with the community, setting up my work, and will also be briefing about the project I am executing.
Do read my first blog about getting to work with CERN-HSF for Google Summer of Code'21 which you may find here.
Table of Contents
1. Interacting with the community
2. About my project
3. Setting up the Development Environment
4. Conclusion
1. Interacting with the community
The first meeting with my mentors was scheduled for the 21st of May, 2021. Me as well as my fellow student developers were integrated with the TMVA Team of ROOT Project, and were directed to sign-up for a CERN Lightweight Account to join the TMVA Channel in Mattermost. CERN uses Mattermost as their instant messaging tool for communications. Mattermost is an open-source, self-hostable online chat service designed as an internal chat for organizations, similar to Slack. The TMVA Team met every Friday to discuss work progress and planned deliverables for the following week. In the first meeting, I met with my mentors for the project along with other GSoC'21 Student Developers and CERN Summer Student Developers working for the TMVA Team of ROOT.
All the GSoC Student Developers working with CERN-HSF were also invited to join a Community Chatroom in Gitter to contact the fellow students and the CERN Administrators. A Welcome Meeting for discussing the community practices and necessary guidelines about working with CERN for the period of GSoC was held on the 3rd of June, 2021 by the CERN-HSF GSoC Team led by the admin Dr. Andrei Gheata, Experimental Nuclear Physicist at CERN.
The Root-Project of CERN, being a data analysis framework typically used in the High-Energy Physics community to handle petabytes of data, consists of many sub-groups, which include TMVA (Toolkit for Multivariate Analysis), RooFIT (Functionality for Modelling and curve-fitting), ROOT-I/O, etc. working for various core features and sub-modules of ROOT. My mentor for the project Lorenzo Moneta also invited all the student developers at the TMVA Team to attend the Weekly meetings of ROOT, where the developers of all the sub-teams of ROOT discussed their work and proceedings, every week on Monday.
2. About my project
ROOT Storage of Deep Learning Models in TMVA
The Toolkit for Multivariate Analysis (TMVA) is a sub-module of ROOT, which provides the Machine Learning environment for training, testing, performance evaluation, and visualization, popularly used in the field of High-energy Physics. Along with core methods supporting ML algorithms like Boosted Decision Trees, Deep Neural Networks, etc., it also has bindings for Python and R, thus interfacing with popular Machine Learning frameworks like Keras, PyTorch, etc. Their training and testing are performed with the use of user-supplied data sets in form of ROOT trees or text files, where each event can have an individual weight. For training, TMVA works using a Factory class object where the required MVA methods are registered and the pre-analysis and pre-processing of the training data is performed to assess basic properties, and subsequently, the training job is called running each MVA method on the provided data. The resulting training details, model configuration, and weights are then written into a .xml file for each of the methods. A lightweight reader class object is used later to read and interpret the .xml file (interfaced by the corresponding MVA method) for inference.
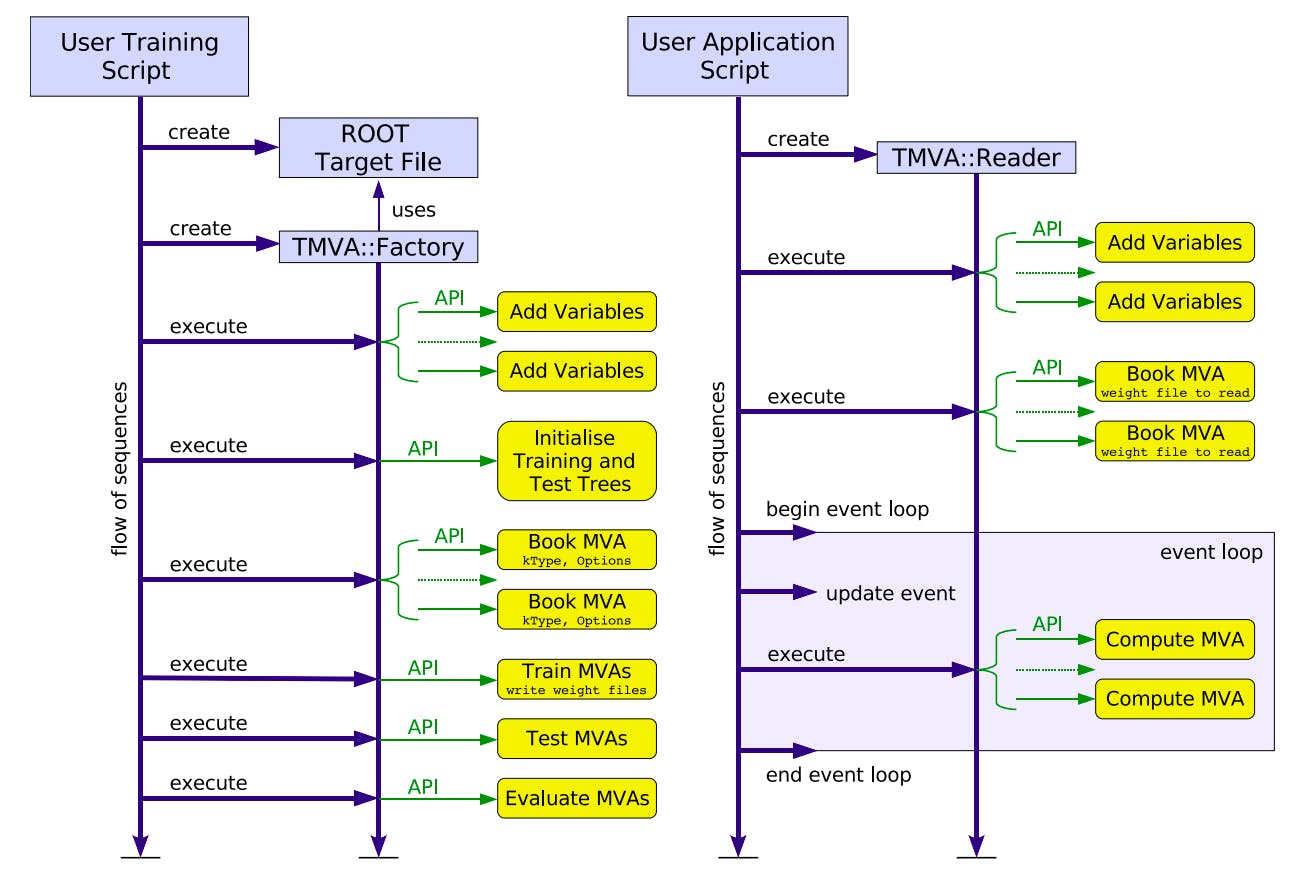
The process of training the methods and successive storing the model data is done by the chain of execution of the following methods:
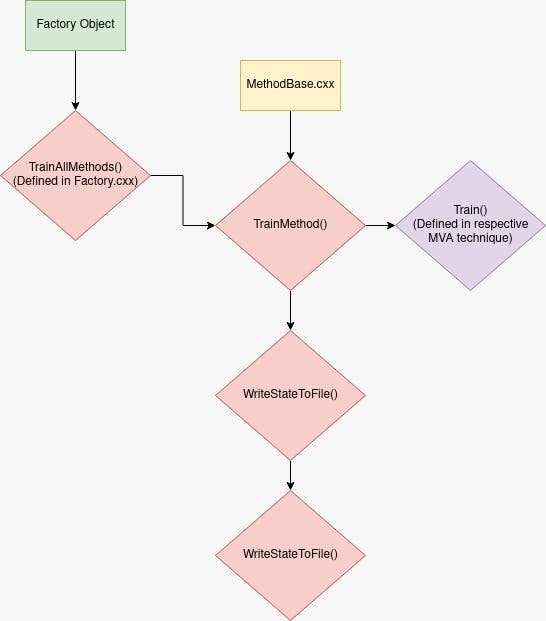
The .xml file stores a lot of information, about both the model, the data used for training, weights, etc.
Properties saved after training
Enter ROOT Files!
Using ROOT, we can write objects and data on a disk into a .root file, thus making them persistent. When reading the file back, the object is reconstructed in memory, for which the ROOT type system requires the memory layout of that object. A .root file is like a UNIX file directory, containing directories and objects in an unlimited number of levels, stored in a machine-independent format. Constructing the .root file uses compression based on the gzip algorithm supporting nine levels of compression. The serialised file is similar to .h5
and .pt files which are used to store trained models in Keras and PyTorch frameworks respectively.
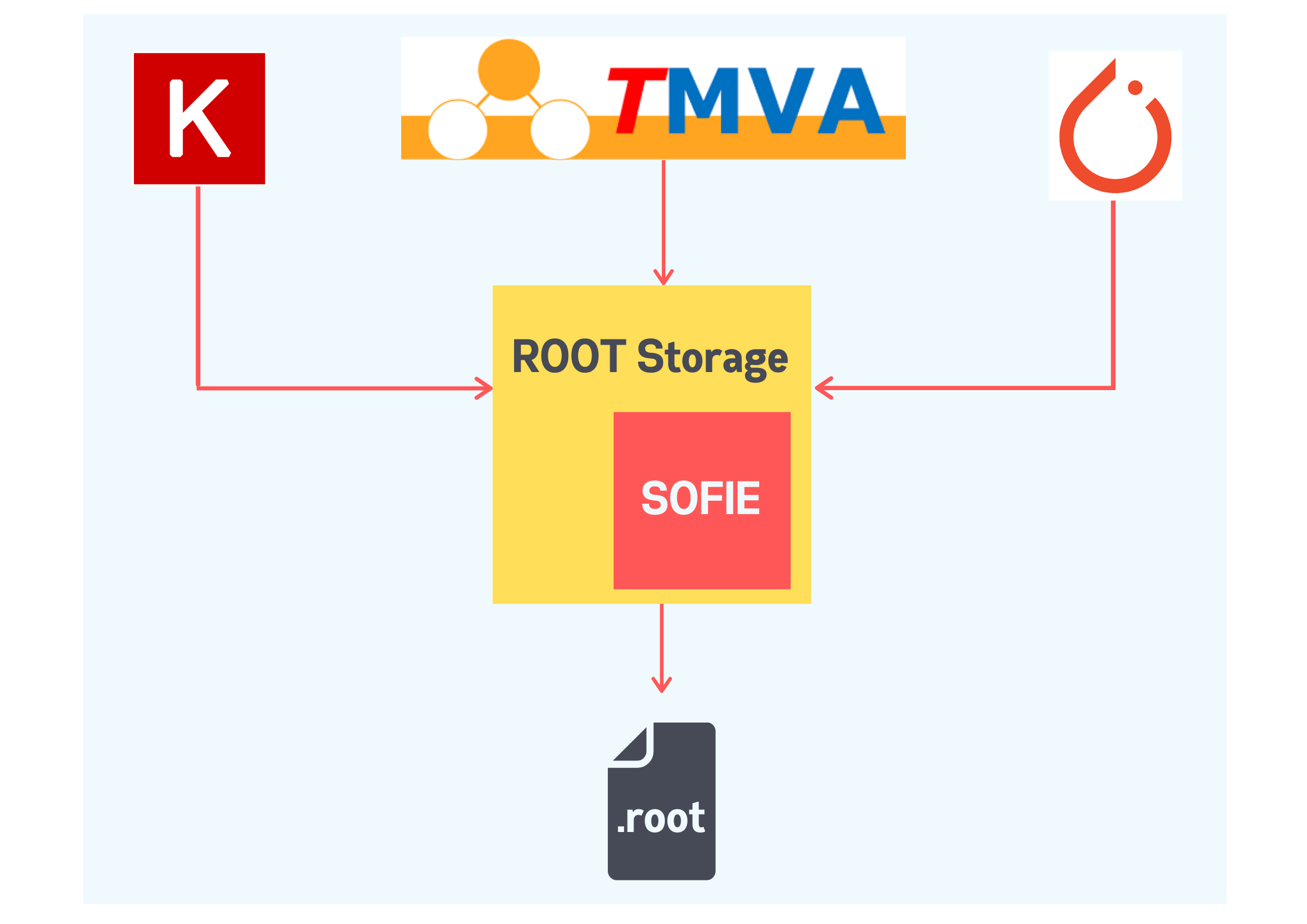
Hey SOFIE!
The TMVA team recently introduced SOFIE (System for Optimized Fast Inference Code Emit), a functionality for inferencing trained models based on the ONNX Standards. SOFIE primarily holds the RModel class which stores the model configuration and weights. It can be used to generate easily invokable C++ functions, for fast inference. SOFIE, currently, also has a parser for translating an ONNX file to store the configuration into the RModel object.
My project aims for developing the functionality to store any trained model in TMVA into ROOT files, for which the trained model is required to be translated into a RModel object, and subsequently to be serialized into a .root file. For a unified standard interface to store any trained model, converters for external frameworks like Keras (.h5) and PyTorch (.pt) are also to be built to store the underlying model architecture and weights into a .root file.
3. Setting up the Development Environment
ROOT uses the CMake build-generator tool as the primary build system. Building the ROOT framework from the source is a strenuous activity, took around 3 hours to build with 4 cores in my local machine. ROOT gets linked with the Python libraries present on the machine, to enable the PyROOT interface, allowing using the Python Bindings, and accessing the Python Interface from C++ using the C-Python API.
SOFIE requires Protobuf>=3.0 for parsing ONNX files into RModel and BLAS or Eigen for the execution of the generated code for inference.
git clone https://github.com/root-project/root.git
mkdir build install && cd build
cmake -DCMAKE_INSTALL_PREFIX=../install -Dtmva-sofie=ON ../root
cmake --build . --target install -j4
source ../install/bin/thisroot.sh
SOFIE needs to be enabled during the build if its usage is required. We may disable other ROOT modules during the build if they are not to be used. The above steps finish the entire build for installing ROOT, and thus after this, we may launch the ROOT framework from CLI using the root command.
The first learning and blocker I faced during this project, was while installing SOFIE with ROOT. I wasn't much familiar with CMake previously, thus beginning this project, I learned about using CMake to build from source code. Installing Protobuf3 was required as it is a pre-requisite to SOFIE, but I faced issues during the process and its configuration. Firstly, I was using Protobuf3.6.1, but the installation threw me some errors, which mainly said, error: ‘class onnx::ModelProto’ has no member named ‘ParseFromIstream’; did you mean ‘ParseFromString’? I asked my mentor Sitong An for help on this, and he advised me to use the latest version of Protobuf, as this error was seen for Protobuf<=3.6, while he worked on tweaking the code of SOFIE to make it compatible fully for versions>=3.

Even after installing Protobuf3.17.2, I was facing issues on configuration, as ROOT was unable to find the latest version. It was facing conflicts with the different versions of Protobuf present in my system. Issues were solved at last, after proper configuration to avoid any conflicts and with a fresh build, I was able to install and successfully use SOFIE.
4. Conclusion
In the three weeks of the Community Bonding period, I learned a lot, especially about CMake and the C-Python API, and dived deep into ROOT and TMVA for having seamless work in the Official Coding period. Interacting with the community at CERN helped me to understand their work culture, project guidelines, and conventions. After a thorough discussion with my mentors, I finalized my working strategy and plan. Following which I began my work on the project.
As of now, while publishing this blog, a substantial amount of the project is finished, and can be found here,
Link to the forked repository: github.com/sanjibansg/root
Link to the prototyped repository: github.com/sanjibansg/TMVAFastInferencePrototype
After the first evaluations this week, a detailed blog will be posted next, which would incorporate my experiences and work-activities in the first half of the official coding period of GSoC'21.
Adiós
Sanjiban
